Development of Moloney Murine Leukemia Virus Reverse Transcriptase Fused with Archaeal DNA-binding Protein Sis7a
Por um escritor misterioso
Descrição

Development of Moloney Murine Leukemia Virus Reverse Transcriptase Fused with Archaeal DNA-binding Protein Sis7a

Ramachandran plot of VEB-1 β-lactamase model from Escherichia coli
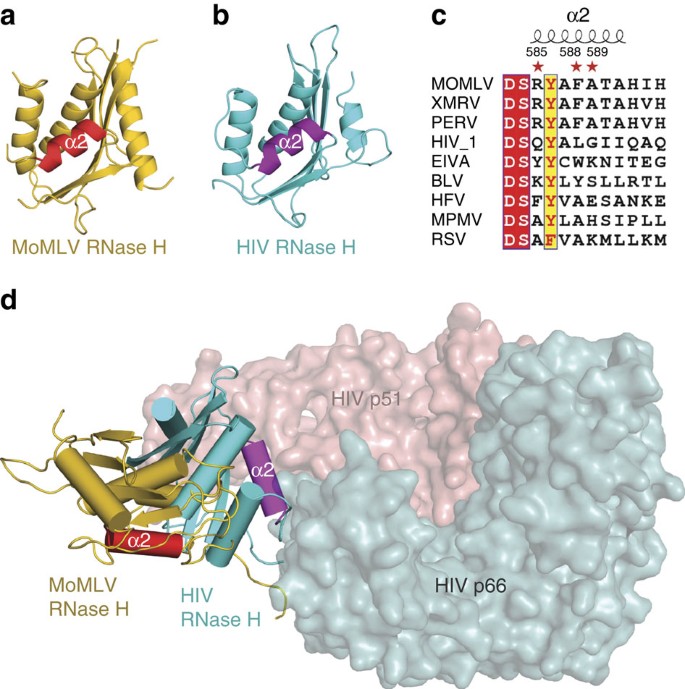
Structural basis of suppression of host translation termination by Moloney Murine Leukemia Virus

Development of Moloney Murine Leukemia Virus Reverse Transcriptase Fused with Archaeal DNA-Binding Protein Sis7a

Processivity analyses of Taq‐based polymerases. Each trace represents

Characterization of a fusion NeqSSB-TaqS DNA polymerase in comparison

Electropherogram traces of Pfu and Pfu-S for the processivity analysis.

Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection
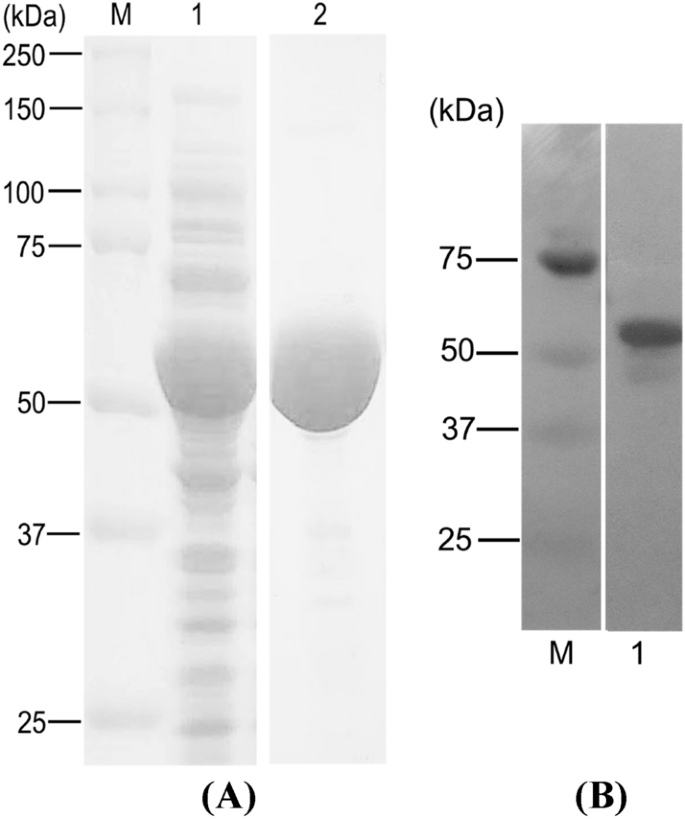
Expression of Codon-Optimized Gene Encoding Murine Moloney Leukemia Virus Reverse Transcriptase in Escherichia coli

The protein homeostasis network in E. coli cells. Protein homeostasis

Amplification efficiency for DNA polymerases depending on the

Reverse Transcriptase of Moloney Murine Leukemia Virus Binds to Eukaryotic Release Factor 1 to Modulate Suppression of Translational Termination: Cell







